The following side project, conducted between my thesis submission and graduation, was aimed to address the mechanistic question that arose during our publication of p53-regulated SESN1 and SESN2 regulate cell proliferation and cell death through control of STAT3 — “What does Sestrin do to influence the activity of STAT3?”
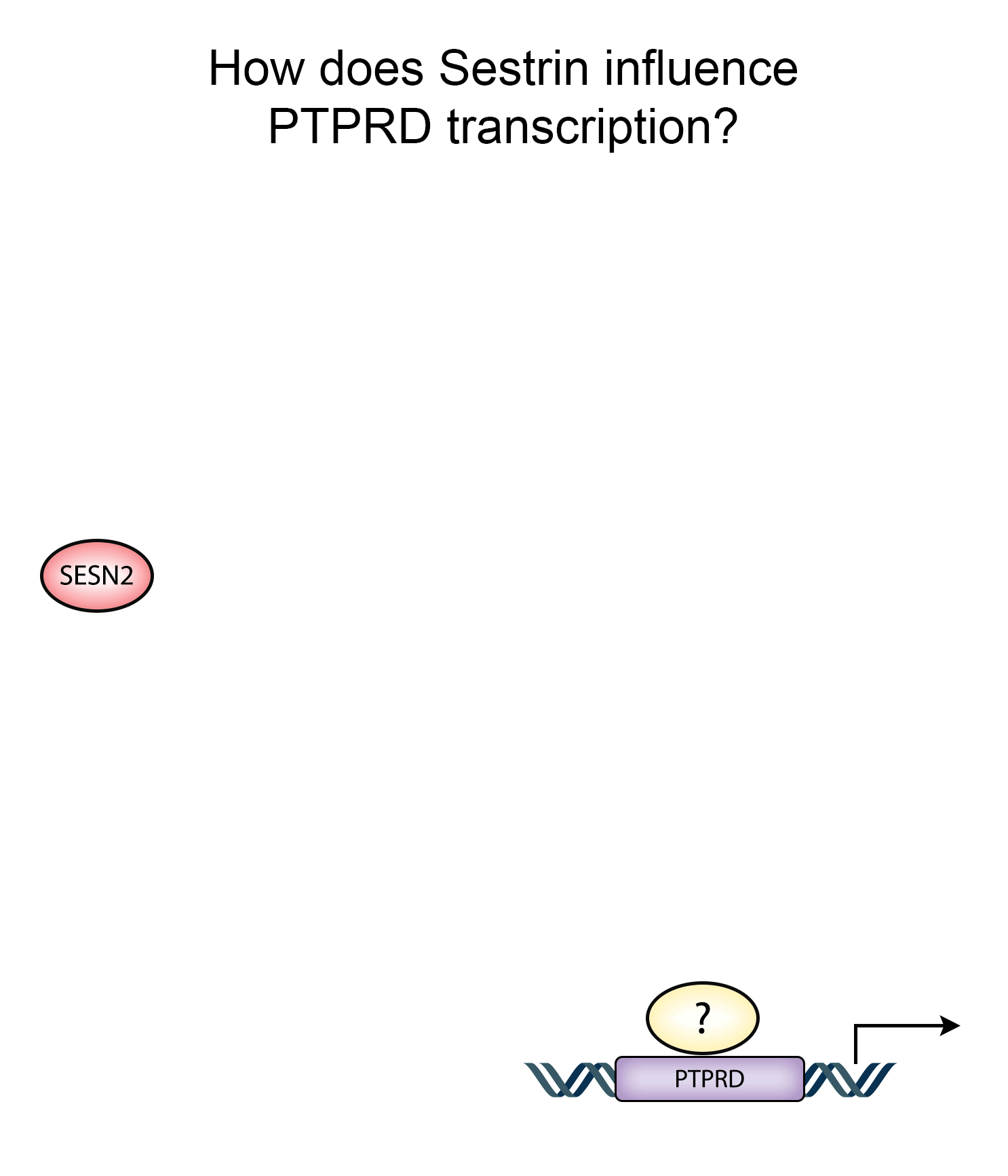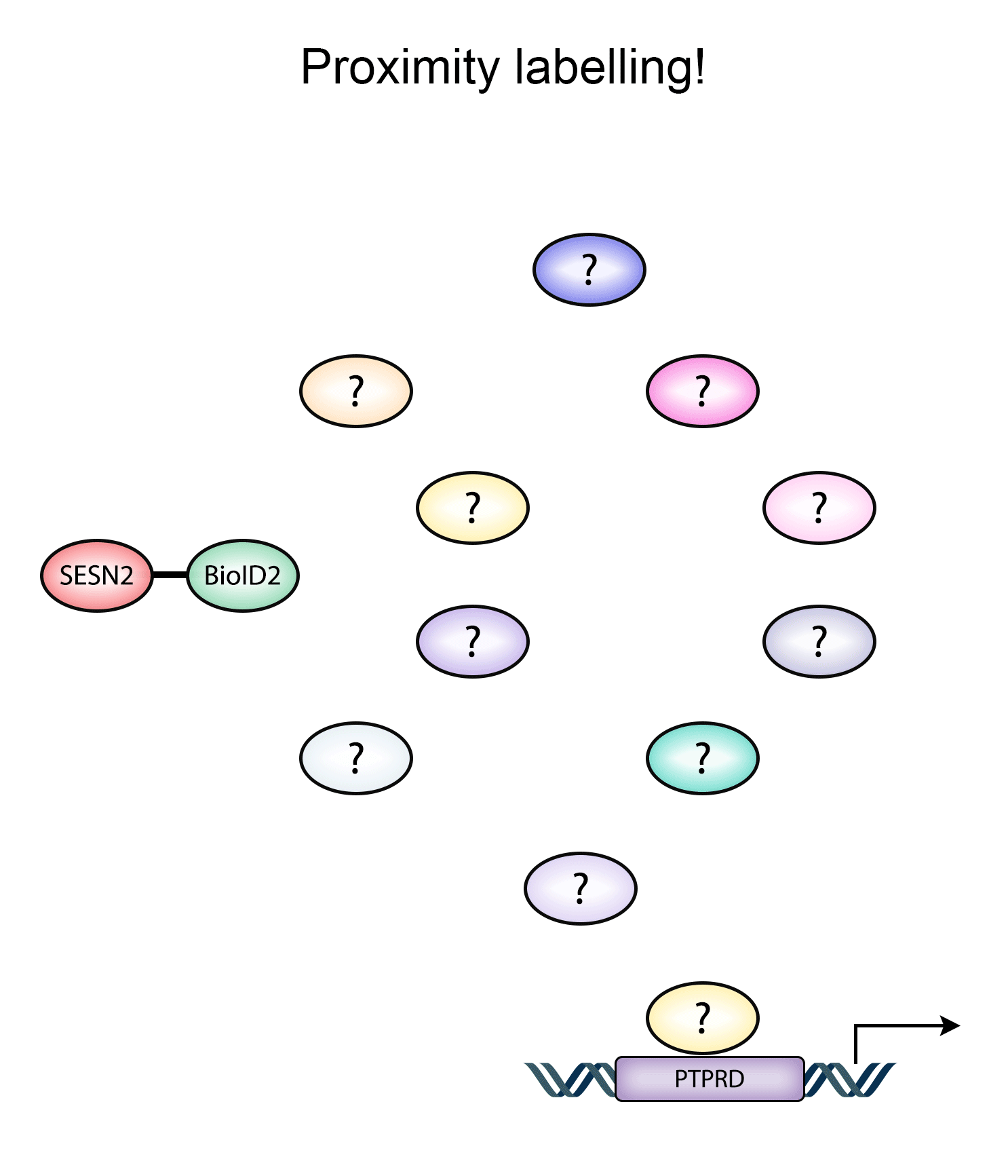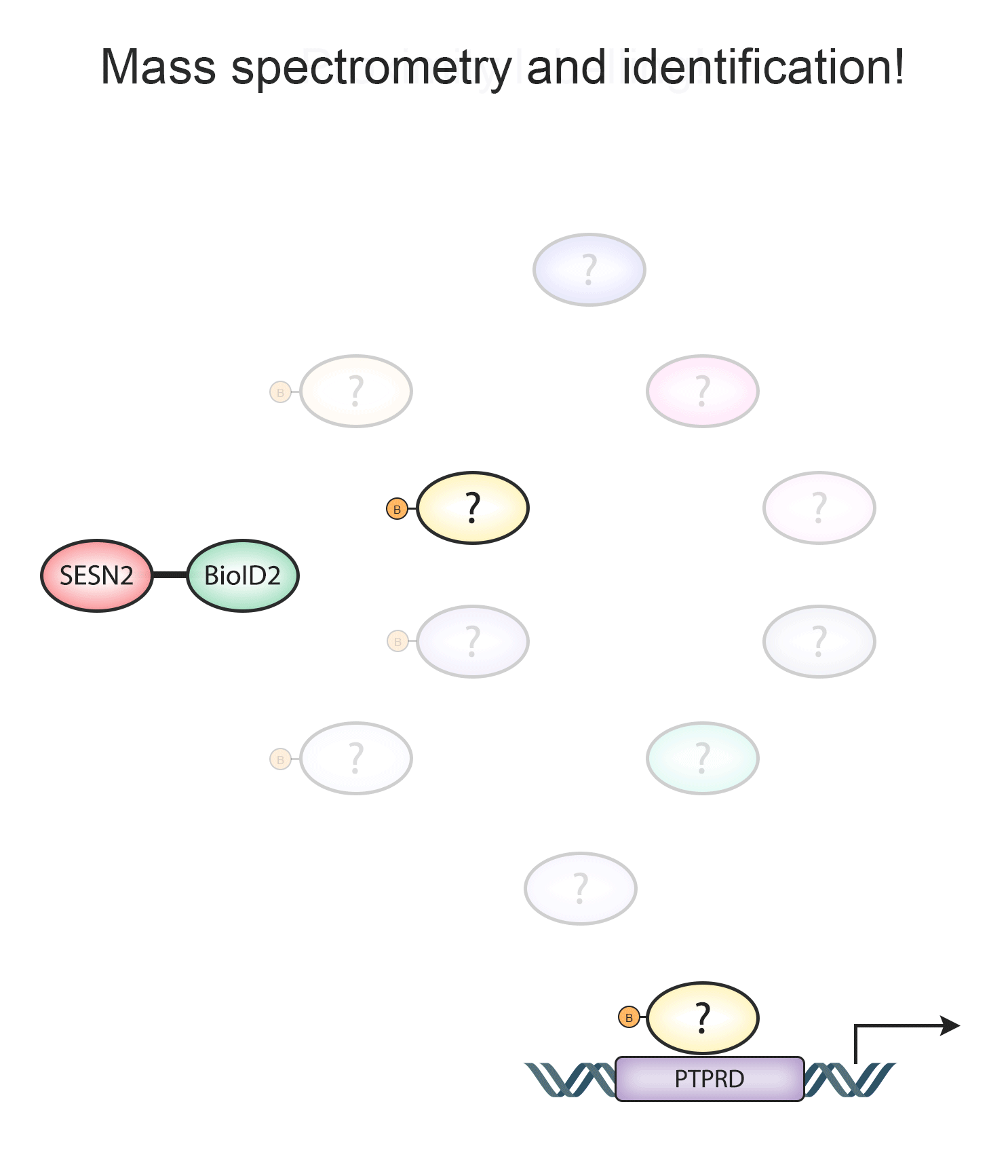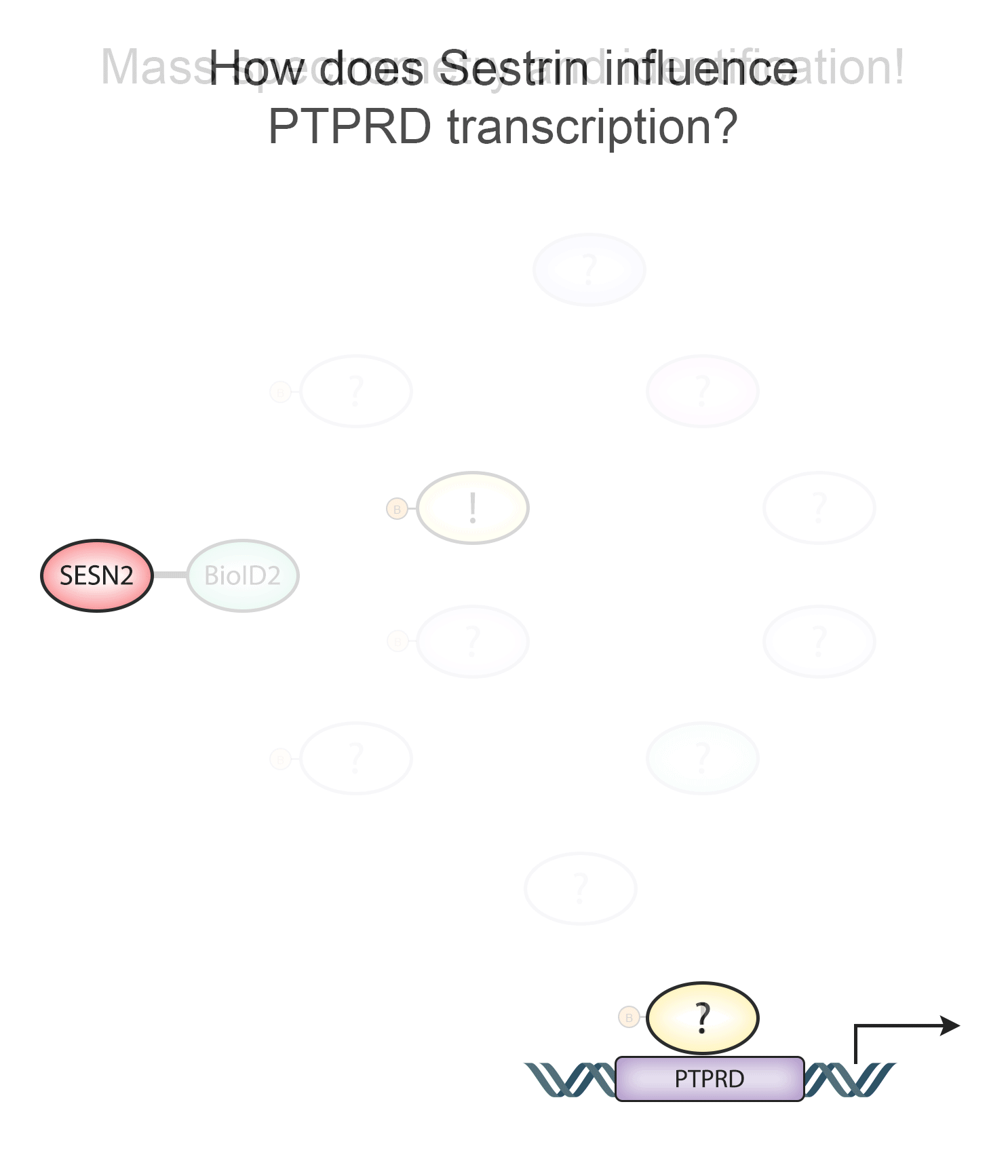
An approach that we were looking to adopt is proximity labelling, where a promiscuous biotin ligase is fused together with a protein of interest, and the resulting biotinylation profiles can be analysed by mass analytical approaches, yielding new datasets and identifying potential interacting partners.
We are currently looking for further funding to conduct a study utilising this construct.
Animated Figure - Using BioID2-SESN2 fusion construct to identify Sestrin interactors
The animated figure displays how BioID2 fused to SESN2 can biotinylate proximal proteins and potentially identify members of other pathways that may not be directly interacting with the Sestrin protein.
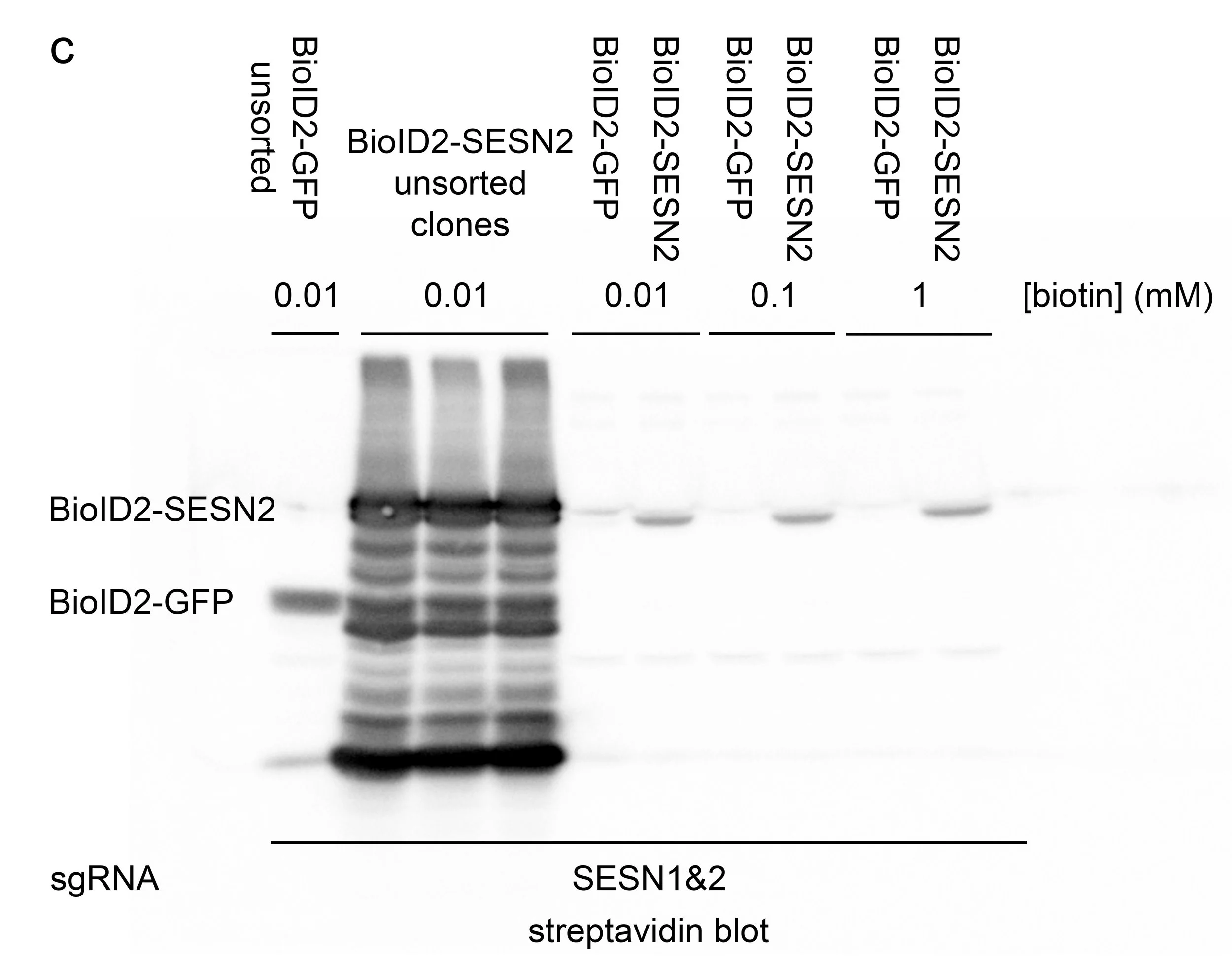
Using molecular cloning, we generated constructs that express BioID2-GFP and BioID2-SESN2. We prepared A549 SESN1&2 knockout cells that stably express both constructs. Furthermore, we thoroughly selected clones of these cells that express a relatively small amount of either construct. Our BioID2-SESN2 express a comparable amount of SESN2 when compared with endogenous wild-type expression (Figure A). Our BioID2-GFP is not detectable by fluorescence microscopy, and we had to use Western Blot to confirm that GFP is expressed in these cells (Figure B). Maintaining low expression of BioID2 constructs is crucial for labelling specificity; however, this often renders biotinylation undetectable by Western blot, requiring the use of more sensitive and costly methods for detection. (Figure C).
Figures - Preparing the BioID2 system in A549 cells
A A glimpse of the selection process for isolating clones with minimal expression of BioID2-SESN2. Cultures with low infection efficiency were subjected to fluorescence-activated cell sorting, in which single cells were dispensed into individual wells of a 96-well plate, allowed to proliferate, and subsequently analysed by Western blotting for SESN2 expression. Clone AA6 exhibited expression levels comparable to the endogenous basal expression of SESN2.
B BioID2-GFP was not detectable by fluorescence microscopy. However, the expression could be visualised using an anti-GFP antibody and detected with an anti-chicken Alexa Fluor 680 secondary antibody, followed by analysis using a LI-COR imaging system.
C Streptavidin-HRP blot showing A549 BioID2 clones before and after fluorescence-activated cell sorting. Notably, the ladder-like pattern of biotinylation observed in the unsorted samples reflects the non-specific biotinylation resulting from high BioID2 expression. Maintaining minimal construct expression facilitates the detection of genuine interaction partners. However, the detection of low levels of biotinylated proteins may require the use of more sensitive techniques, such as mass spectrometry.
We have now confirmed that the BioID2-SESN2 construct is capable of binding its GATOR2 partner WDR24.
Future studies will utilise these cell lines, and the metaphorical baton of this project is now in the hands of a future research assistant or PhD student in our lab.
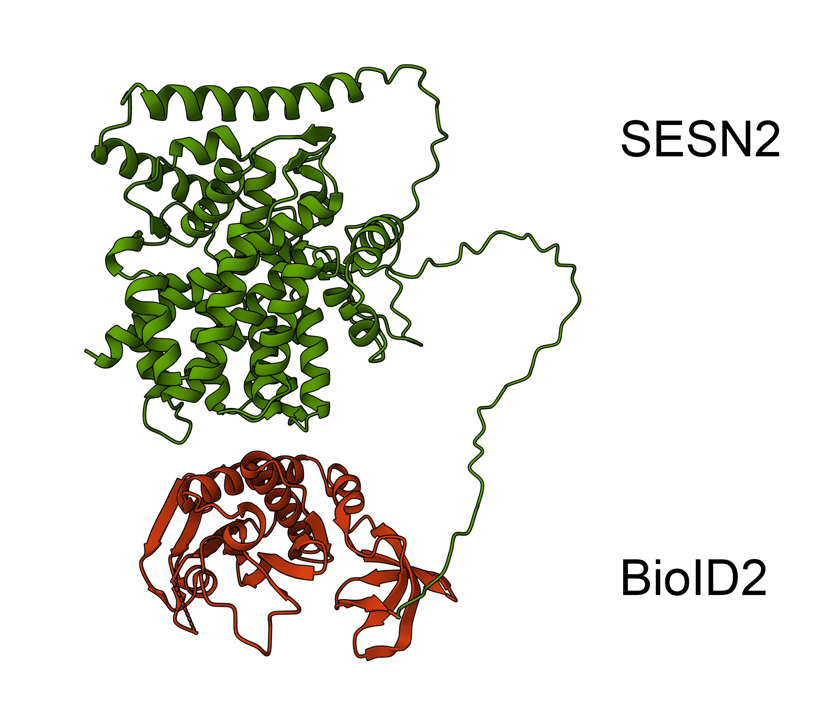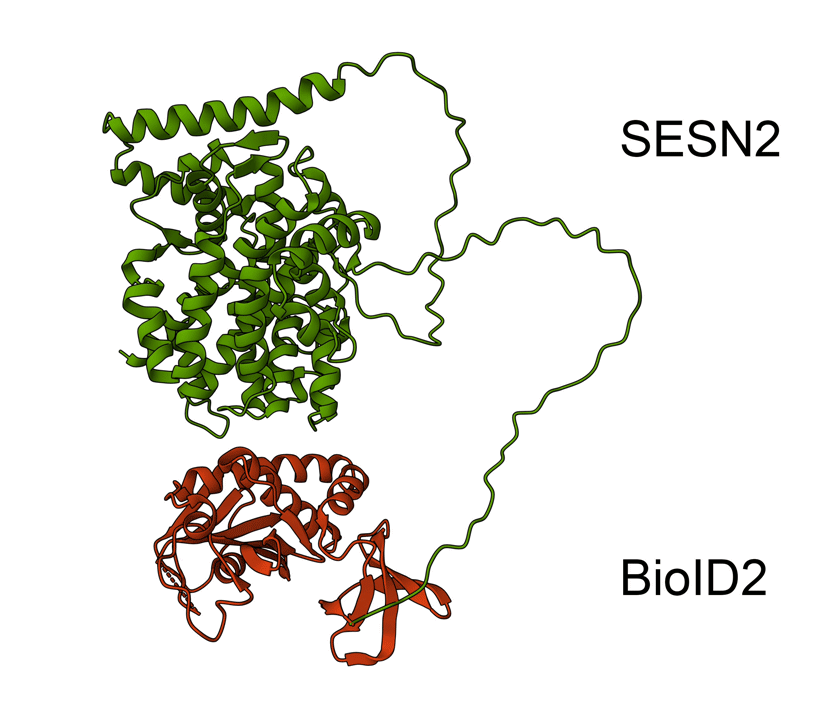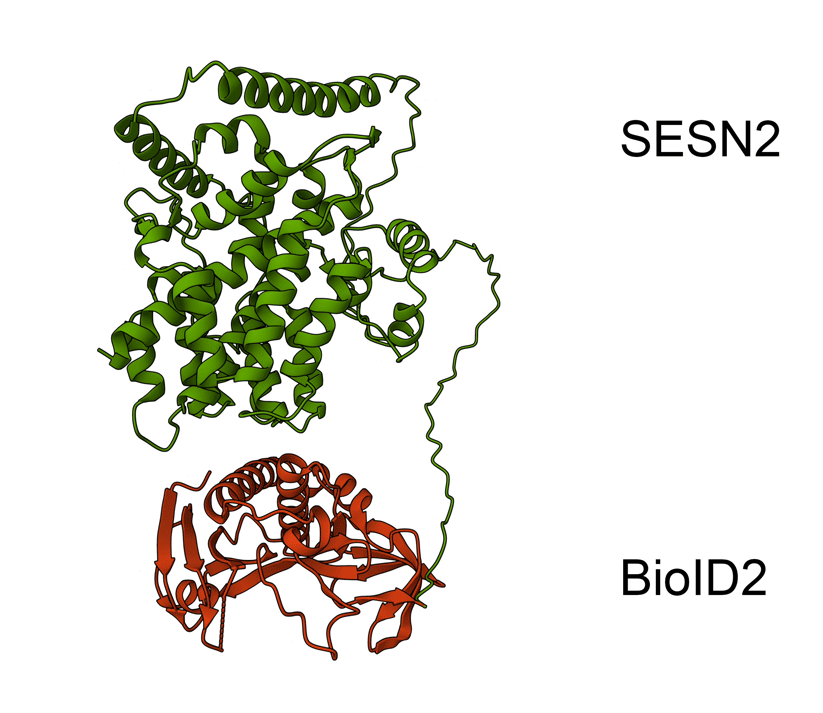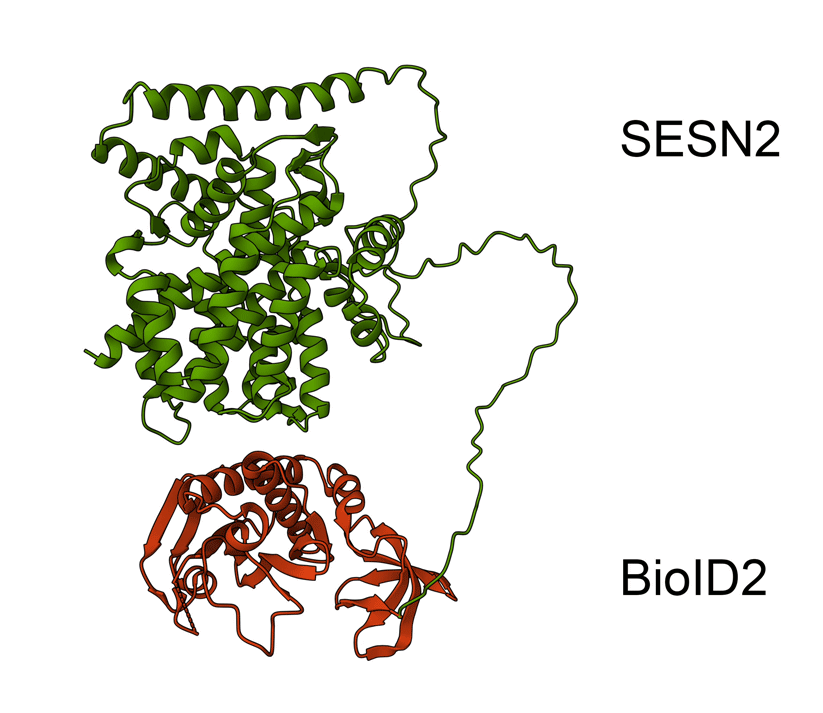
Animated Figure - the hypothetical structure of the BioID2-SESN2 construct
We fused BioID2 onto the N-term of SESN2 to generate a BioID2-SESN2 construct. The Alphafold structure of SESN2 (AF-P58004-F1-v4) and the crystal structure of Biotin Ligase from Aquifex aeolicus (2EAY) were fused in PyMOL using the bond command, for a hypothetical visualisation. The N-term of SESN2 has a long disordered tail, which likely means that our fused BioID2 does not hinder SESN2 and can freely engage in proximal labelling.
Green - SESN2. Red - BioID2.
Here is an example showing the importance of maintaining low BioID2 construct expression.
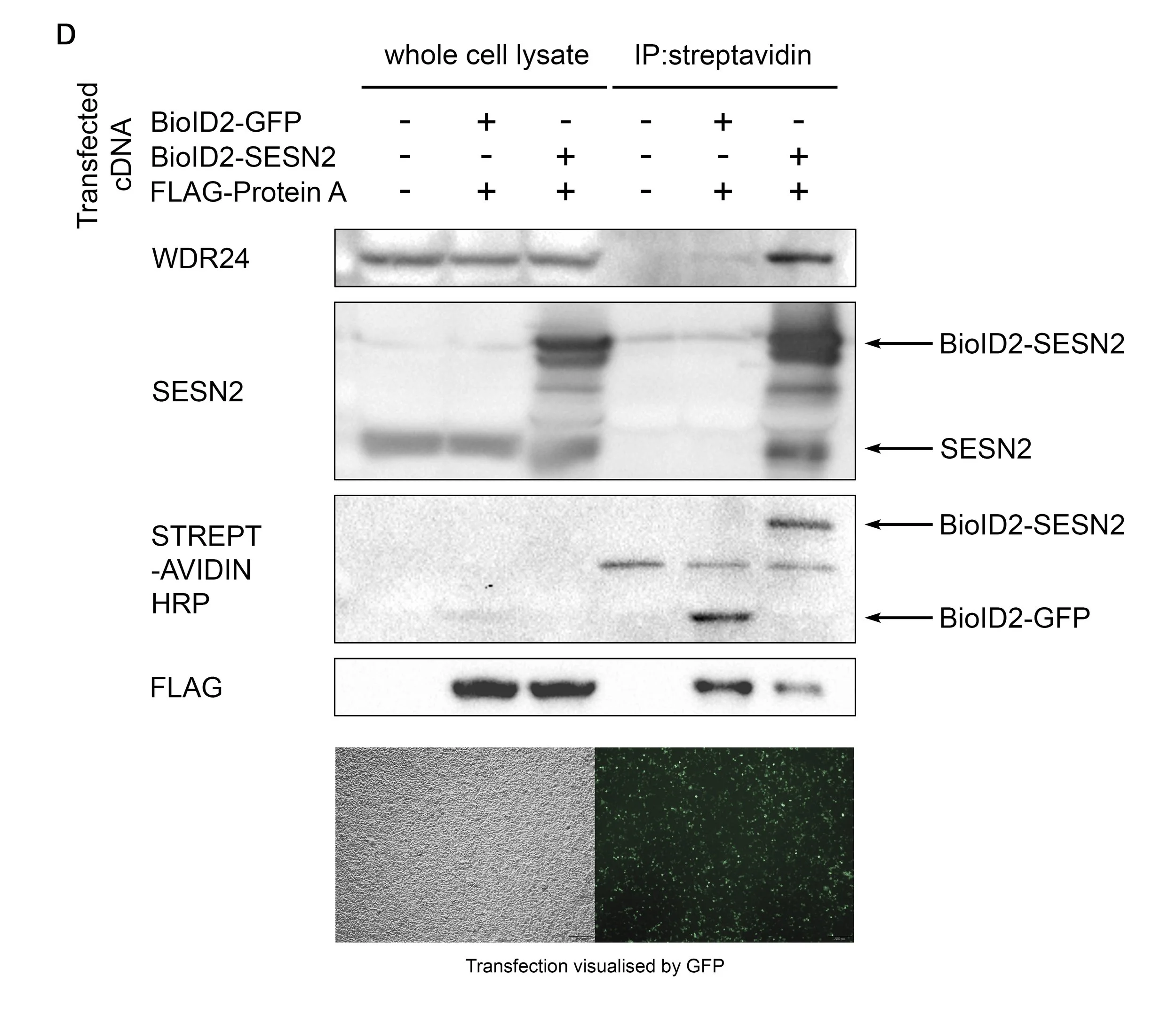
To assess the potential interaction between SESN2 and a putative binding partner, here referred to as Protein A, a preliminary proximity-labelling experiment was conducted using the BioID2 system. Wild-type HEK293T cells were transiently transfected with either BioID2-GFP or BioID2-SESN2 constructs, together with FLAG-tagged Protein A. Following incubation in the presence of 5 µM biotin, cells were lysed, and the lysates were subjected to streptavidin-mediated pulldown to isolate biotinylated proteins (Figure D).
When using transient transfection, it is inherently difficult to regulate expression levels. Here, we observed that excessive expression of either the BioID2-GFP construct or Protein A resulted in non-specific biotinylation. A significant portion of Protein A was pulled down in the BioID2-GFP sample.
Therefore, transient overexpression is not suitable for evaluating the proximity between SESN2 and Protein A. It is important to utilise carefully selected clones of a stable cell line with low and uniform BioID2-SESN2 expression, ensuring that biotinylation is minimal to preserve specificity.
Figure - The pulldown of proteins according to their biotinylation status
D Wild-type 293T cells were transfected with either BioID2-GFP or BioID2-SESN2, together with FLAG-Protein A. Cells were incubated with 5 µM biotin overnight, then lysed and subjected to streptavidin bead pulldown. As seen in the FLAG blot, BioID2-GFP biotinylated a large portion of Protein A, likely due to its overexpression, as the SESN2 target WDR24 was only marginally detected in the immunoprecipitate. Therefore, the proximity between SESN2 and Protein A cannot be reliably studied using a simple transfection.