While conducting a large sequence alignment analysis for Sestrins, we noted that several putative Sestrin-like sequences are found across the Trypanosoma genus.
We found an entry for a putative Sestrin-like protein in Trypanoma brucei (strain brucei TREU927):
On TriTrypDb (Tb927.10.13470), also found on NCBI (XP_827831.1) and UniProt (Q388Q1).
Our colleagues in the next lab over work with this strain of Trypanosoma brucei.
Thus, we dubbed this gene TbSESN and attempted to isolate it.
We received a spare pellet of centrifuged trypanosomes from our colleagues, and we isolated their RNA. Following reverse transcription, we performed a PCR to see whether this gene is found as an expressed RNA in the Trypanosoma brucei. The PCR on cDNA showed a positive result and we designed flanking primers to clone the gene.
After a round of cloning, we isolated the gene into the pLU plasmid.
Figures - The cloning of Trypanosomal Sestrin
A The first sight of TbSESN after PCR amplification for cloning. B Ligation into pLU plasmid (vector). C Restriction digest of the amplified ligation products.
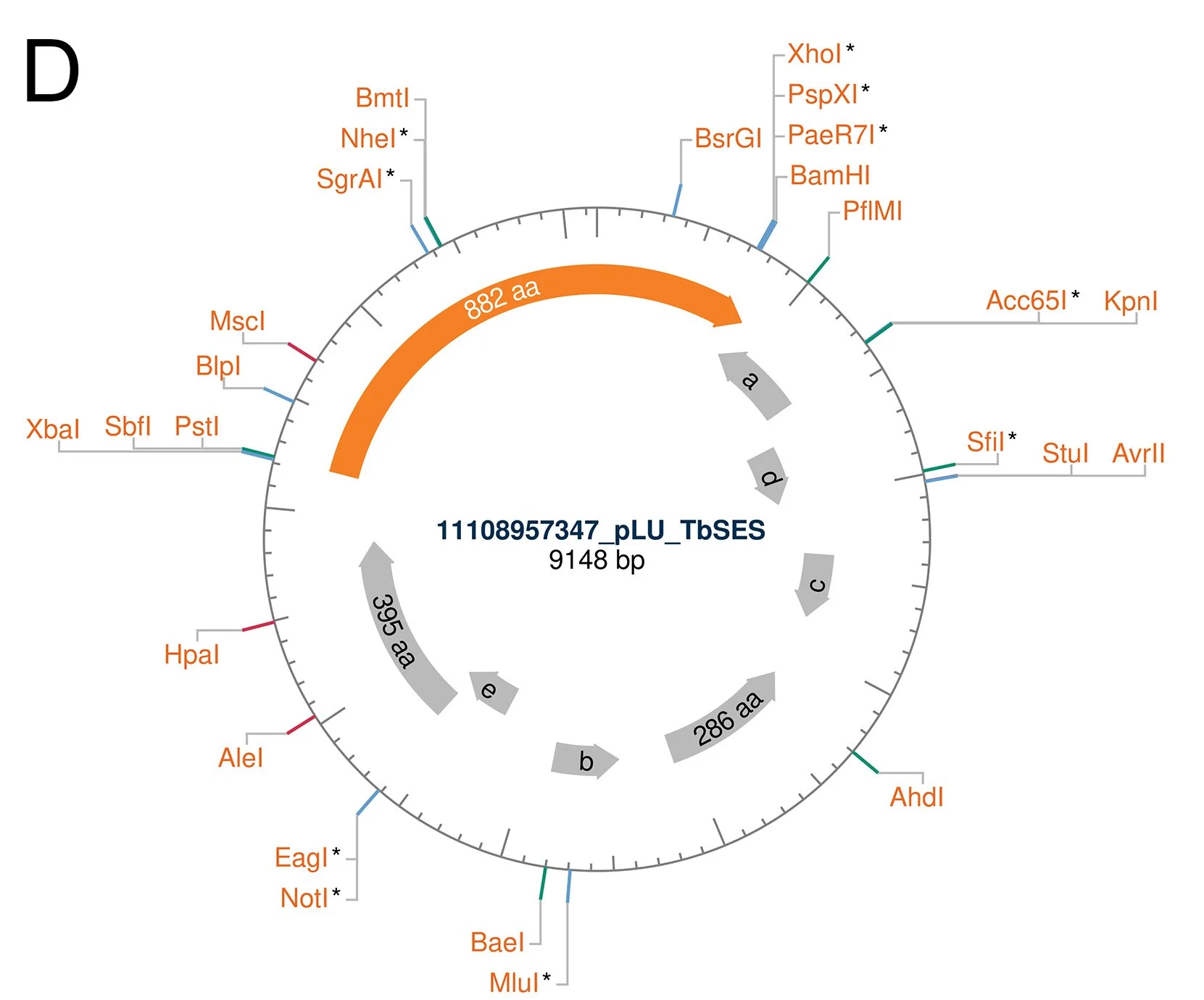
The plasmid with the isolated gene was sent for sequencing, and it returned displaying an open reading frame (ORF) that matched in size with our cloning target (Figure D).
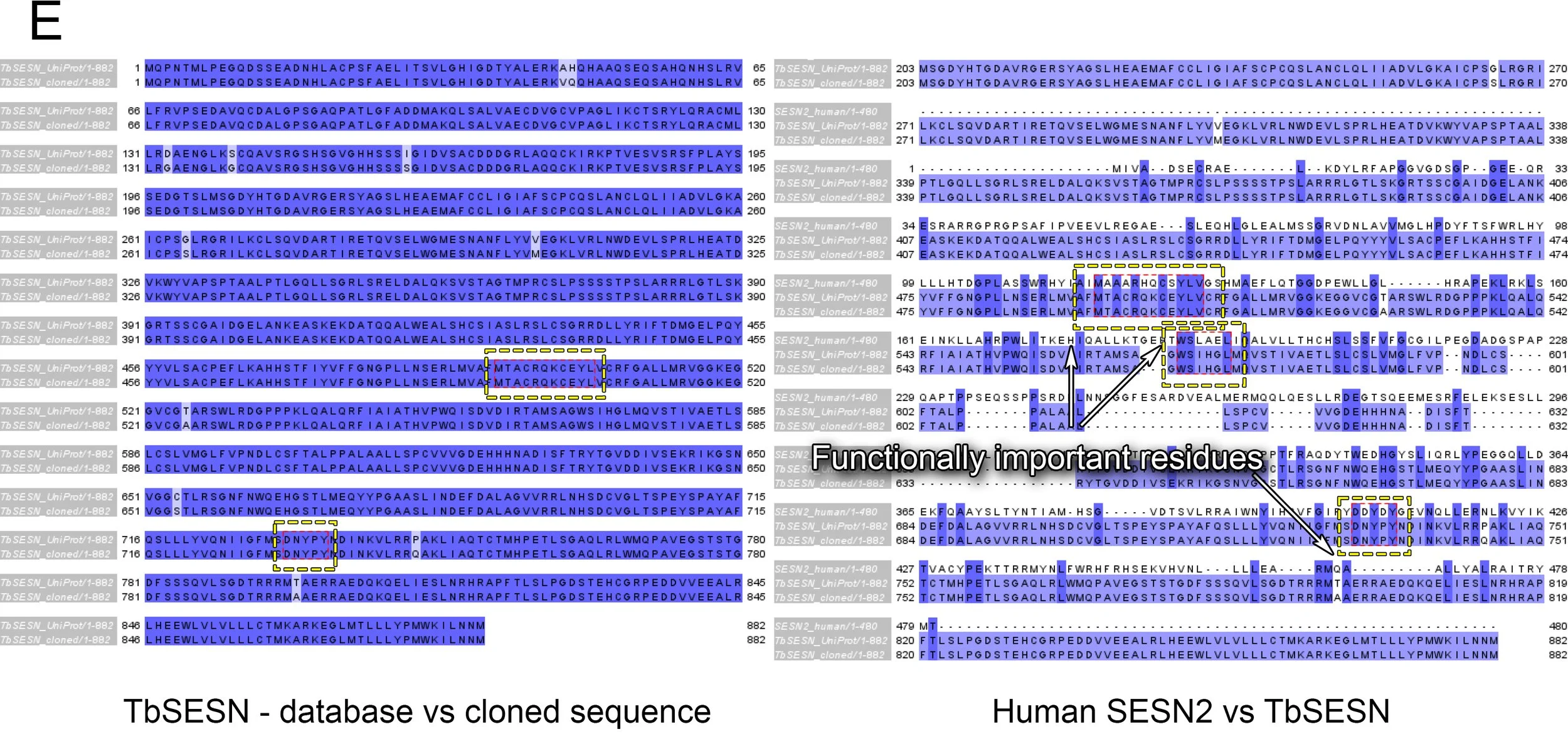
Sequence comparison via Jalview (Figure E below) displayed that our cloned gene matched closely with the UniProt database entry.
Moreover, we could see close conservation of functionally important sites when comparing the human SESN2 with the cloned TbSESN:
DDYDY - the motif responsible for GATOR2 binding is present as DNYPY, containing the functionally important aspartic acid (DNYPY) and two tyrosines (DNYPY). In most Trypanosoma species, two of the aspartic acids are found (DDYxY). However, in Trypanosoma brucei, this site is instead a polar uncharged arginine residue (DNYPY).
WS - the N-term pair of residues responsible for GATOR2 binding, is also present in TbSESN.
CxYL - the antioxidative motif of SESN2 is also present in TbSESN.
BLAST shows a strong conservation of the DDYDY, WS, and CxYL motifs in the Trypanosoma genus.
Figures - The sequence of the cloned TbSESN
D The sequence of the plasmid with the cloned ORF visualised by the NEBcutter 3.0 tool.
E The sequences of human SESN2 and the database entry for putative trypanosomal Sestrin compared with the cloned ORF by ClustalOmega alignment, visualised by Jalview. The amino acid sequence encoded by the ORF was generated using the Expasy Translate tool.
The sequencing of the cloned plasmid shows an ORF that encodes an 882 amino acid protein. The translated sequence generated by Expasy Translate showed little difference between the database and the cloned gene. Moreover, the comparison with the human SESN2 showed that functionally important residues are conserved.
Based on the primary sequence, the residues known for their functions in the human Sestrin are also present in the Trypanosoma equivalent. This insight would be interesting to experiment with, to see how many of the Sestrin functions the trypanosomal Sestrin displays.
Furthermore, clear equivalents of WDR24 & SEH1L (GATOR2 proteins that Sestrin binds) could not be found in Trypanosoma, raising the question of what, if anything, Sestrin interacts with in this organism. If the binding partners are significantly different from GATOR2, this could serve as a model to study the evolution of the Sestrin-GATOR2 binding, and the emergence of mTORC1 regulatory mechanisms.
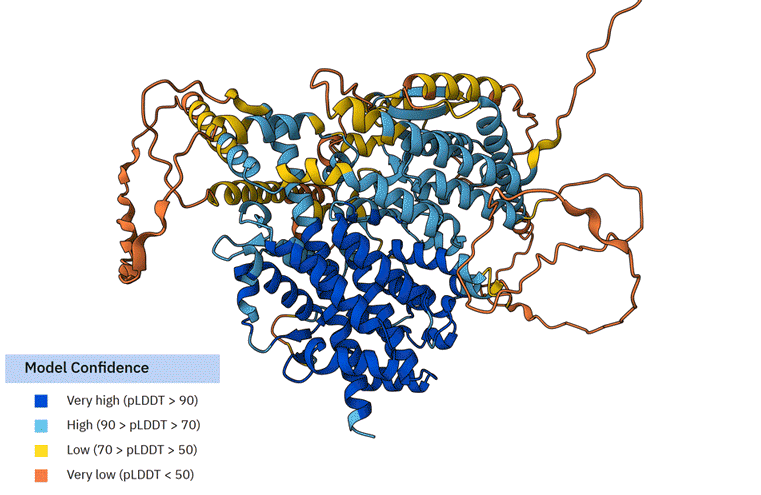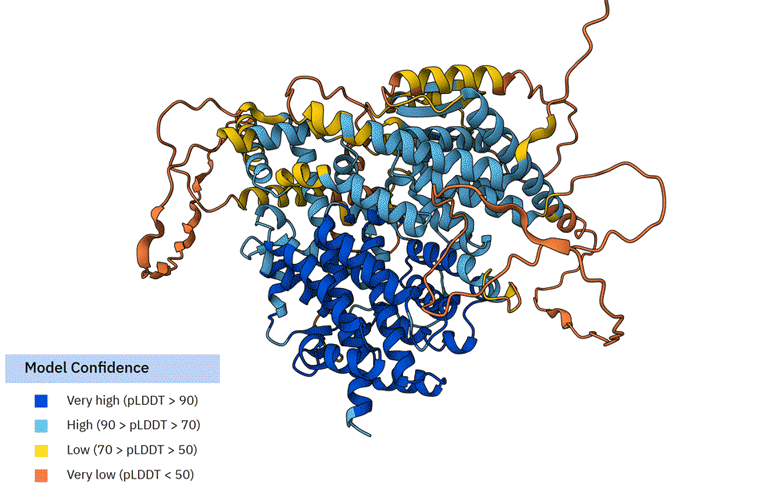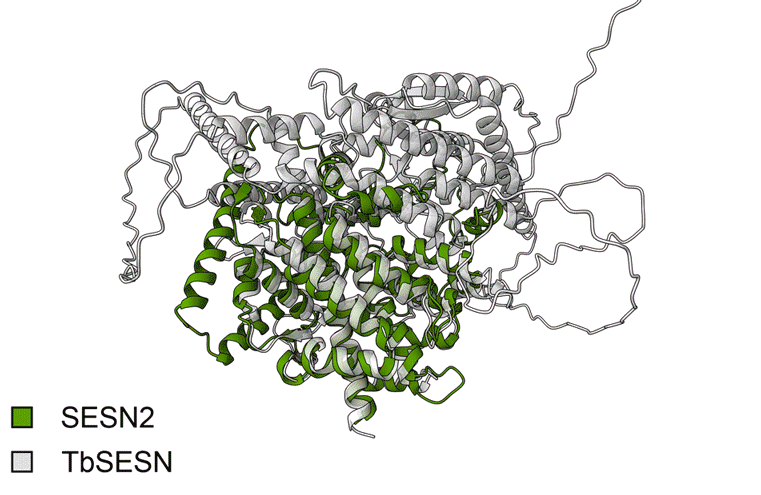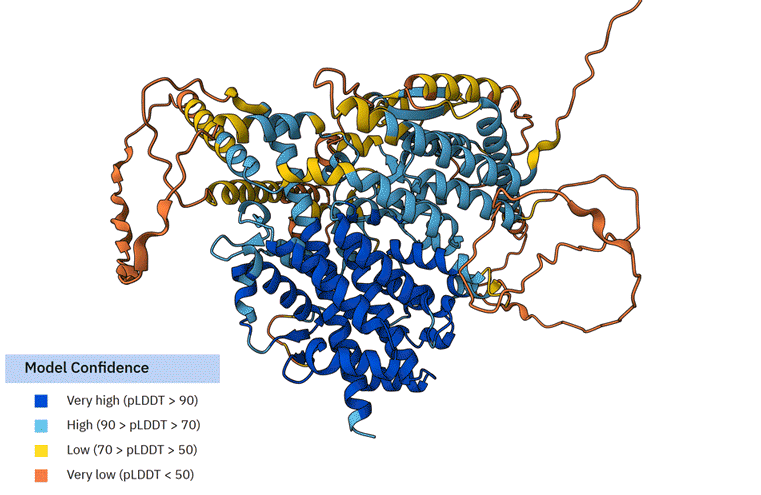
We also compared the similarities of TbSESN to the human SESN2 via the AlphaFold structural database.
The average AlphaFold model score of TbSESN suggests that it is a poorly constructed model (Average pLDDT 64.03 (Low)). The portion of TbSESN that is resolved most accurately (Very high (pLDDT > 90)) is the part of the structure that resembles human SESN2, likely reflecting the AlphaFold model bias towards available resolved structures.
We overlapped the SESN2 and TbSESN structures using PyMOL (Animated Figure F). The structural overlap of TbSESN and SESN2 shows a close fit (RMSD = 2.192). The core SESN2 helices that form its signature structure overlap almost perfectly with TbSESN. However, it is important to note that these are based on AlphaFold models, which are biased towards the human SESN2 structure, the only currently resolved Sestrin structure.
Animated Figure - The overlap of SESN2 and TbSESN
F First animation: The Alphafold model of TbSESN (AF-Q388Q1-F1-v6). Predicted structure is shown with model confidence values indicated by the colour scale. Second animation: The Alphafold model of TbSESN (grey) is superimposed on the Alphafold model of human SESN2 (AF-P58004-F1-v6; green).
Representatives of the Trypanosoma all possess a much longer N-terminus (approximately additional 450 residues compared to SESN2), which is poorly predicted in the AlphaFold model. A brief sequence alignment analysis showed that the N-term varies among the Trypanosoma species more significantly than the core Sestrin-like structure. Only a single motif within this region - YSSEDG - shows conservation among the Trypanosoma species. This motif is within the ‘Very low (pLDDT <50)’ prediction zone. It is therefore difficult to postulate its function or role. Nonetheless, the fact that all trypanosomal Sestrins have an ~450 amino acid N-terminal extension strongly suggests that that region may serve an important functional purpose.
In summary, due to the similarities in structure and functional sites, the trypanosomal Sestrin can be hypothesised to serve similar functions that the mammalian Sestrin serves, while potentially incorporating an additional function unique to Trypanosoma through its N-term extension.
Subsequently, we successfully performed another round of cloning and added a FLAG tag to the N-terminus of TbSESN. The FLAG tag allowed us to perform a couple of checks, such as checking if the construct expresses in 293T cells, and other things. We now possess a plasmid that is ready for the induction of TbSESN in the Trypanosoma brucei (Figures G-I).
The study of TbSESN overexpression, of its effect on the trypanosomal cell, and identification of its binding partners, can reveal the functions and purpose of this protein in trypanosomes. Furthermore, designing shRNA and other methods of studying this protein can yield additional insights as to what its functions are at baseline expression.
The project is currently ready to be taken over by a future student in the laboratory and may pave for a productive collaboration with our Trypanosoma colleagues.
Figures - Attachment of FLAG to TbSESN
G The first sight of FLAG-TbSESN after PCR amplification for cloning. H Ligation of the FLAG-TbSESN PCR product into pLU plasmid (vector). The vector was dephosphorylated using FastAP, therefore not showing non-specific ligation products. I Western blot showing the expression of FLAG-TbSESN in SESN2-knockout 293T cells. GFP and human SESN2 were used as controls.
In addition to generating an exogenous expression construct for TbSESN, we have also explored the possibility of studying its endogenous expression in Trypanosoma brucei.
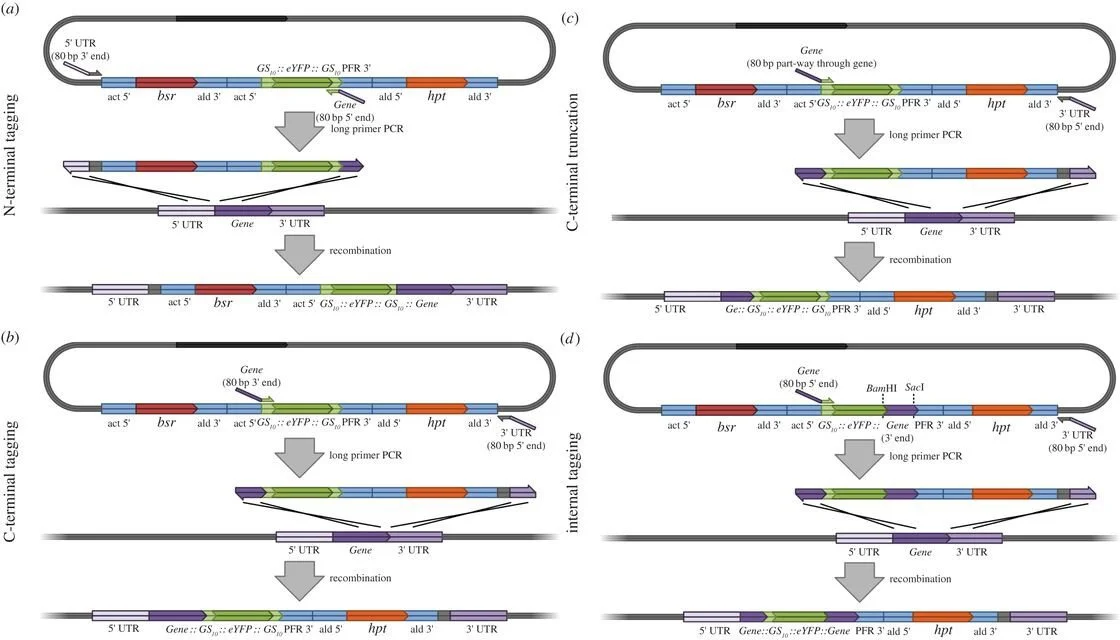
The pPOT plasmid system provides a convenient way for endogenously tagging genes in T. brucei. This approach requires the design of particular 100 base pair (bp) primers. Of these, 80 bp correspond to a sequence homologous to the genomic region spanning the 5′ untranslated region (UTR) or the start codon of the target gene. The remaining 20 bp of the primer are used as annealing sequences for the pPOT plasmid.
Using these primers, a fragment can be amplified from the pPOT plasmid by PCR and introduced into the parasite. Homologous recombination then integrates the fragment in front of the endogenous locus of the gene. This allows the gene to be fused with tags such as YFP or Ty1, enabling the study of endogenously expressed gene using confocal imaging and western blotting, respectively.
More can be read about the following method here:
Figure retrieved from [1]
“Figure 2. Long primer PCR tagging and deletion mutagenesis using pPOTv4. (a) N-terminal tagging. (b) C-terminal tagging. (c) Creating a C-terminal deletion mutant. (d) Internal tagging towards the N-terminal end of the gene (using a modified pPOTv4 template).”